Tony K.Y. Lim, Ph.D.

Tony K.Y. Lim, Ph.D.
RNA Therapeutics, Neuroscience, and Pharmacology

I am an interdisciplinary scientist focused on developing next-generation RNA therapeutics for neurological diseases.
I have engineered an ‘immune-evasive’ self-amplifying RNA platform that inherently reduces the immune-related side effects of this technology.
My future goals are to translate this platform to clinical use and demonstrate its effectiveness in treating conditions such as chronic pain and autoimmunity.
RESEARCH

The molecule used in modern COVID-19 vaccines—messenger RNA (mRNA)—holds significant promise as a therapeutic platform for gene expression. However, its broader application is restricted by its transient nature: once inside cells, mRNA typically degrades within days, limiting the duration of its effect.
To achieve long-lasting therapeutic gene expression, researchers and clinicians often use viral vectors—engineered viruses that deliver genetic material into cells. While these vectors can provide more sustained effects, they also carry important safety risks. For example, viral vectors may unintentionally integrate their genetic material into a person’s DNA, which can potentially lead to cancer. The immune system might also react strongly to the vectors or to treated cells, causing inflammation or tissue damage. Additionally, once the genetic material is delivered, it cannot be easily removed, which makes risk management challenging.
My research seeks to establish a therapeutic platform that balances durability with controllability—outlasting mRNA without the prolonged, less manageable persistence of viral vectors.
To accomplish this, I work with self-amplifying RNA (saRNA), a specialized form of RNA capable of replicating within cells to sustain protein production over time. However, the replication process of conventional saRNA produces double-stranded RNA, a molecular pattern recognized by the immune system as a viral infection. This results in an immune response that can inhibit protein production and may even lead to cell death.

My work addresses this fundamental challenge by pioneering an ‘immune-evasive’ saRNA platform that intrinsically silences the immune alarms it would normally trigger. By engineering the saRNA to co-express a suite of inhibitors that disarm the cell’s ability to detect it, the saRNA can replicate without provoking a harmful immune response. In mouse cells relevant to joint disease, I have demonstrated that this approach enables long-lasting, high-level protein expression with with minimal damage to the cells and reduced inflammatory mediator release.
In contrast, conventional saRNA triggers immune responses that halt protein production, kill cells, and cause inflammation. While medications that suppress the immune system can reduce these reactions, they complicate treatment and increase the risk of infections and other unwanted side-effects. Unlike conventional saRNA, the immune-evasive saRNA platform operates without the need for external immunosuppressants. I also demonstrated that a small-molecule antiviral can act as an ‘off-switch’: at low concentrations it temporarily suppresses gene expression, while at higher concentrations it terminates expression altogether. This level of control provides flexibility during treatment and ensures expression can be halted once therapeutic goals are achieved.
This proof-of-concept study in mouse cells highlights the promise of the immune-evasive saRNA platform to provide durable, controllable gene expression—bridging the therapeutic gap between the transience of mRNA and the persistence of viral vectors.
My future work will focus on advancing this platform toward clinical use. The first step is to understand and overcome immune barriers caused by effector immune cells, which can attack treated cells and limit how long the saRNA remains in the body. These efforts will lead to improved immune-evasive saRNA designs that enable repeated dosing. At the same time, I will adapt the system for human cells by applying computational design to develop inhibitors that specifically target human immune proteins. Ultimately, the goal is to use this platform to develop new treatments, such as turning a patient’s own cells into “bio-factories” that continuously produce peptide therapeutics to help manage chronic diseases such as chronic pain and autoimmune disorders.
EXPERTISE
-
Self amplifying RNA engineering
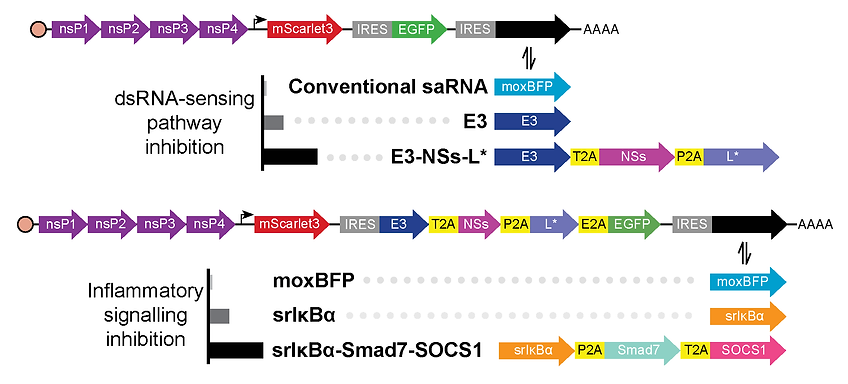
Design of self-amplifying RNA constructs that intrinsically inhibit diverse double-stranded RNA sensing and inflammatory signaling pathways using proteins expressed via cap-independent translation.
Learn more about this approach for therapeutic gene expression here:
-
Microplate assay development and image-based analysis
BioTracker
EGFP
mScarlet3
Mock Transfection
Native saRNA
E3
E3-NSs-L*
moxBFP
srIκBα
srIκBα-Smad7-SOCS1
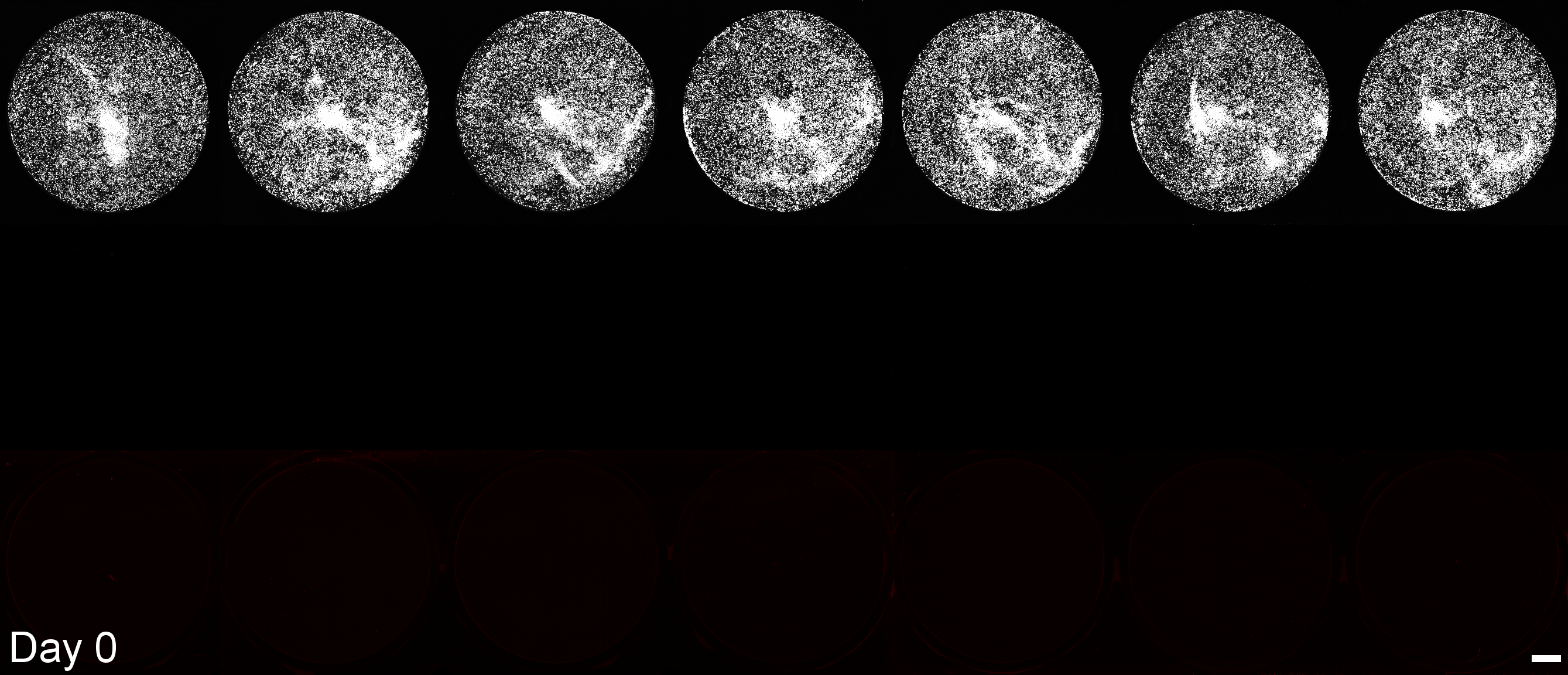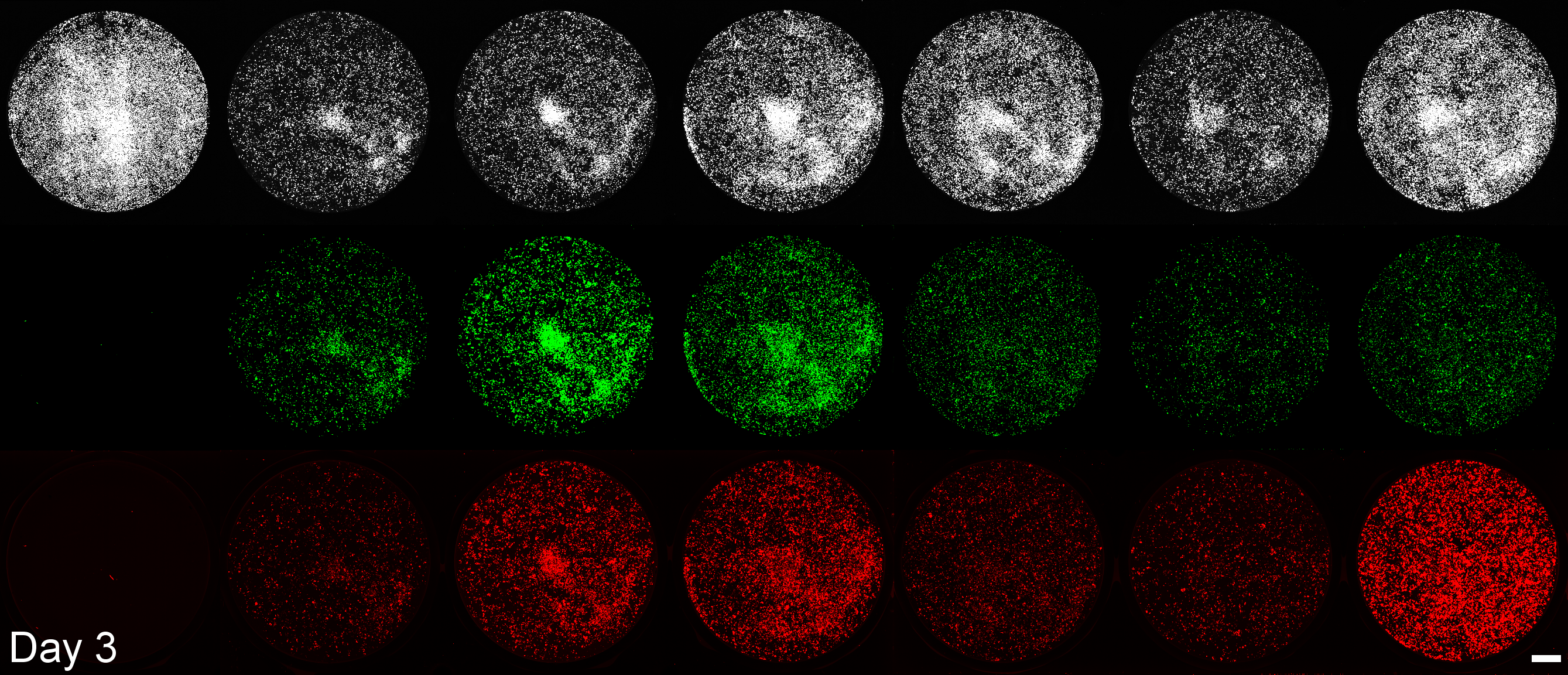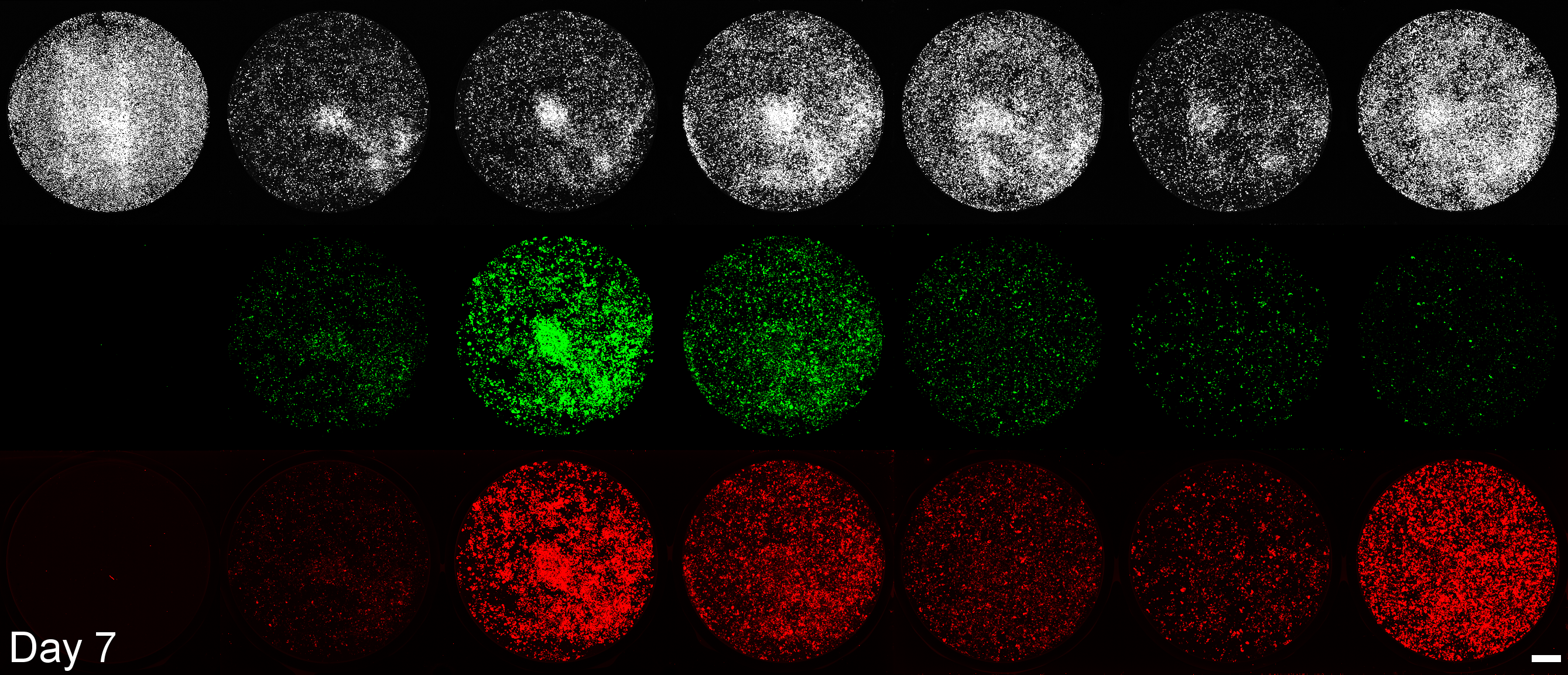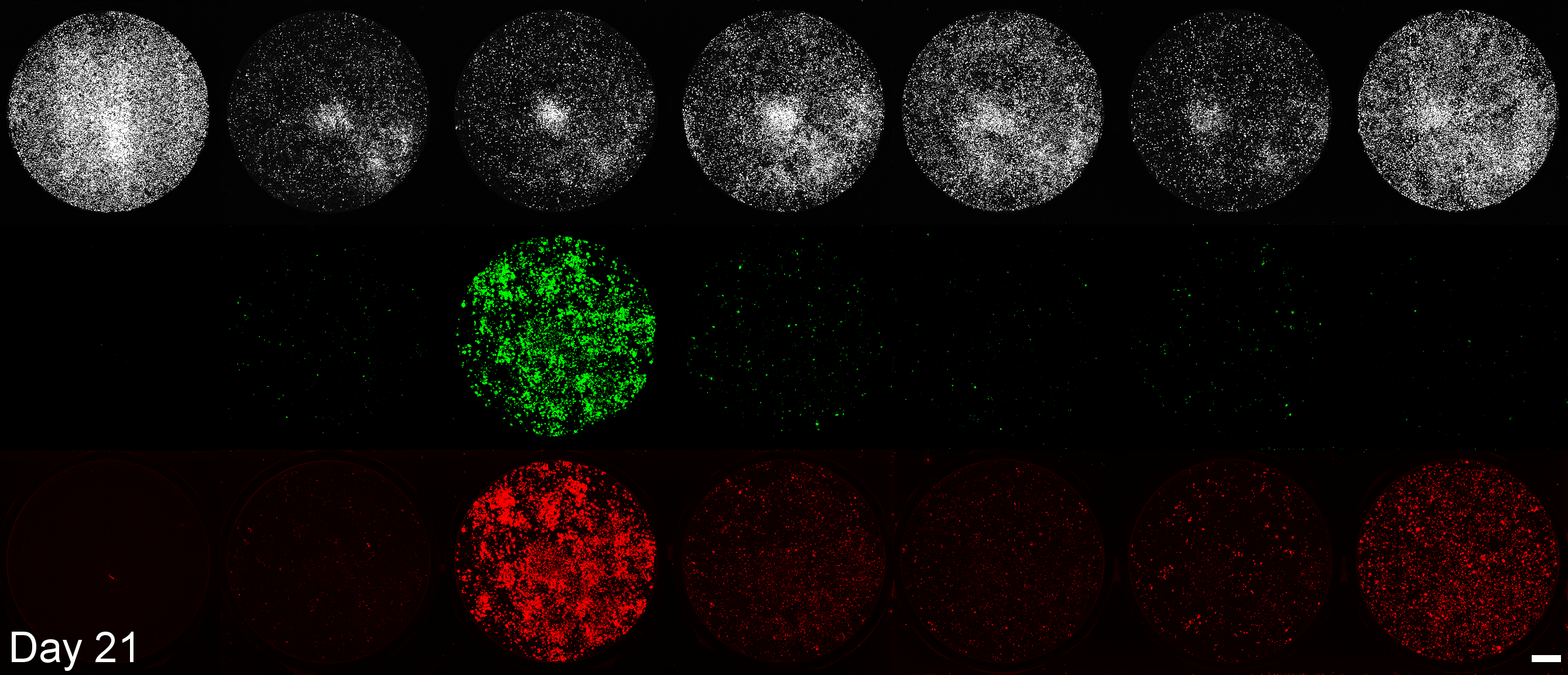
Development of a microplate assay enabling simultaneous, longitudinal monitoring of cell number alongside both cap-independent and cap-dependent transgene expression.
(BioTracker = cell number, EGFP = cap-independent transgene expression, mScarlet3 = cap-dependent transgene expression)
This assay uses the Odyssey M plate imager. Spectral unmixing software for EGFP and mScarlet3 was coded by Dr. Larissa Ferguson and can be found here:
-
2-photon intravital microscopy

Microglial cells phagocytosing neurons (neurophagy) in the developing brain. Timestamp=HH:MM

Microglial cell (red) trogocytosing a pH-stable GFP-labelled axon (green). The colocalization of green and red is shown as white.
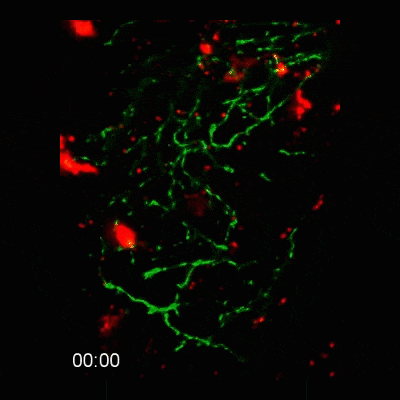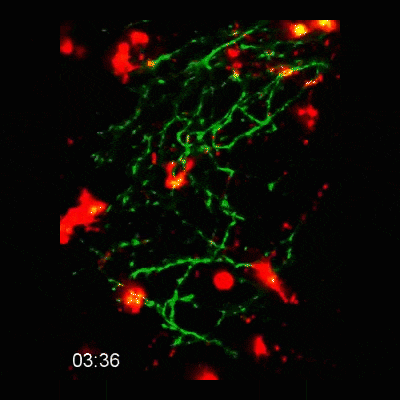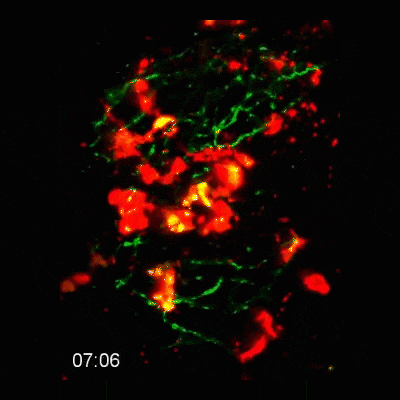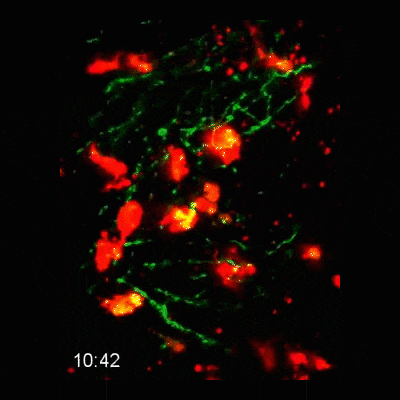
Microglial cells (red) responding to a laser irradiation injury. Timestamp=HH:MM
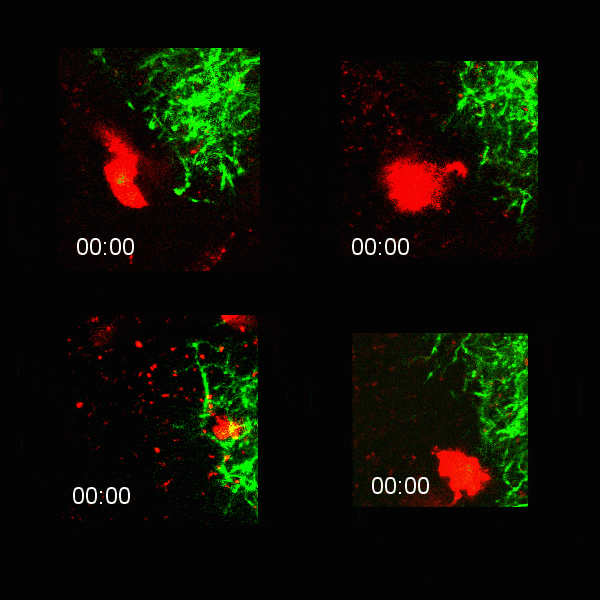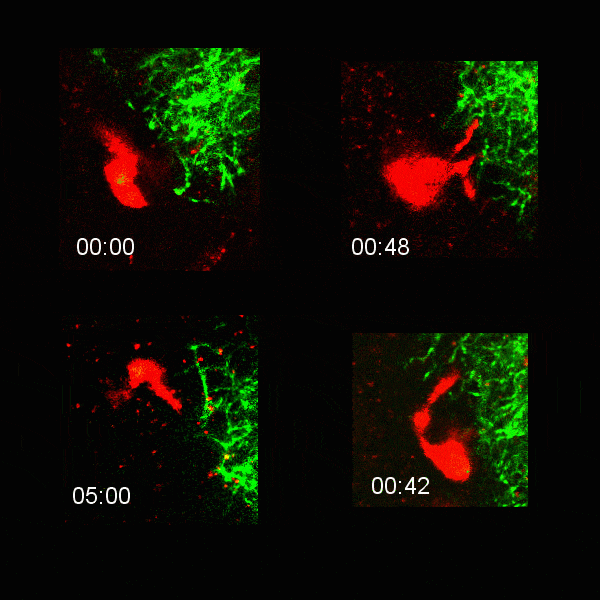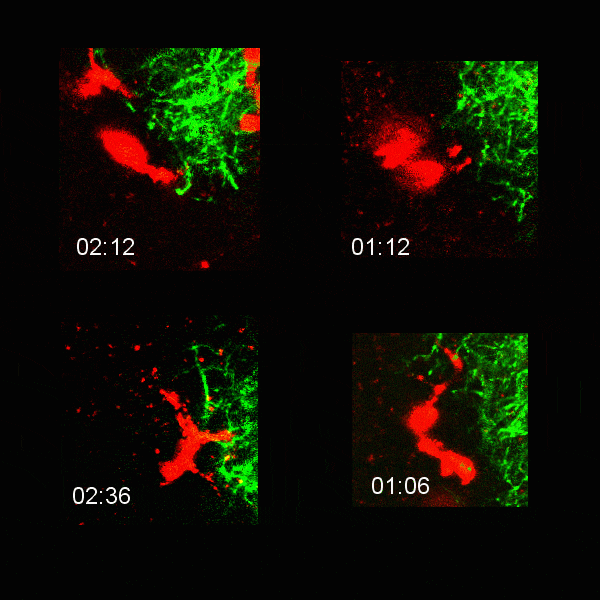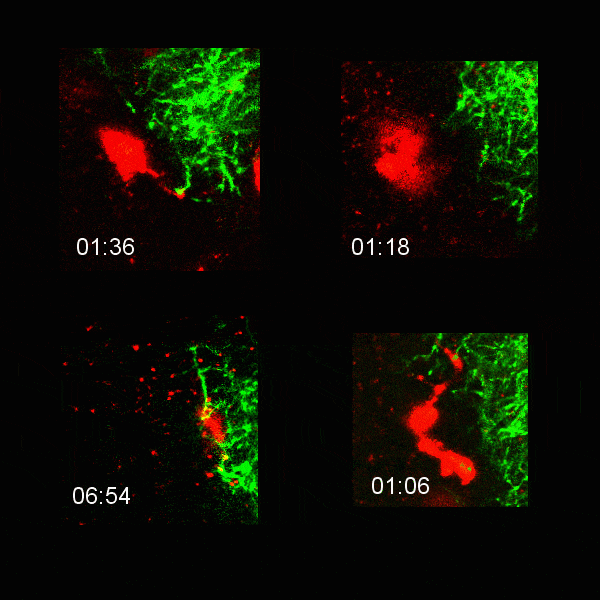
Microglial cells (red) extending processes to contact axons (green). Timestamp=HH:MM

Left: Microglial cells (red) surveil the brain in the presence of an eGFP-labelled axon (green).
Right: Tracking of the microglial cells on the left panel in 3D.
Find out more about this work by checking out the article, published in eLife: https://elifesciences.org/articles/62167
-
Behavioural assay development and video analysis

Simultaneous presentation of looming stimuli and recording of behavioural responses using Python.
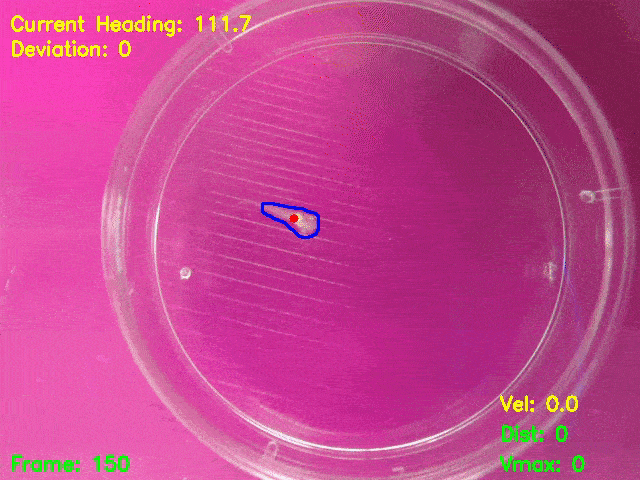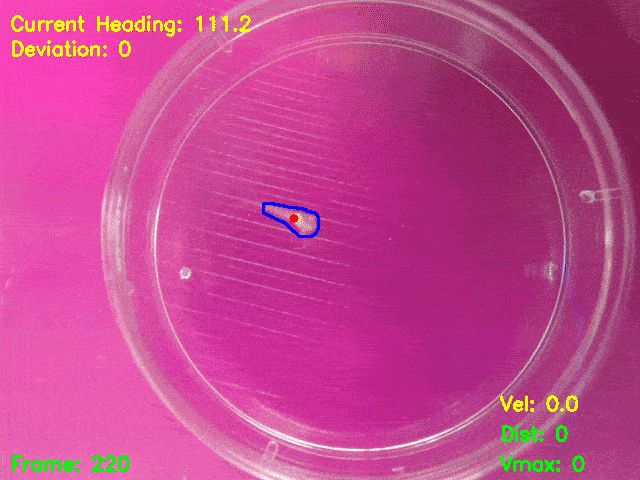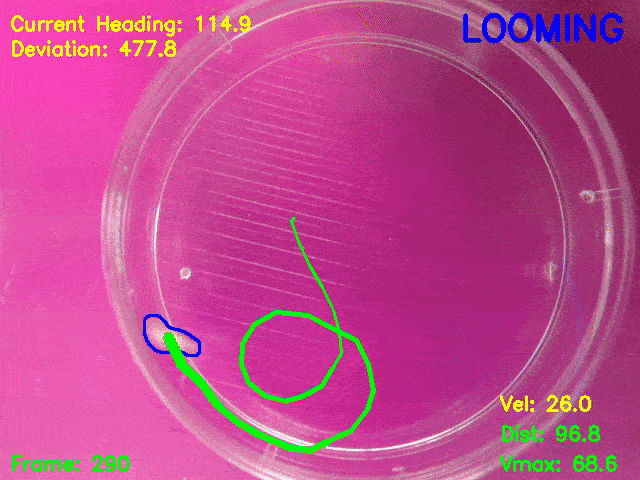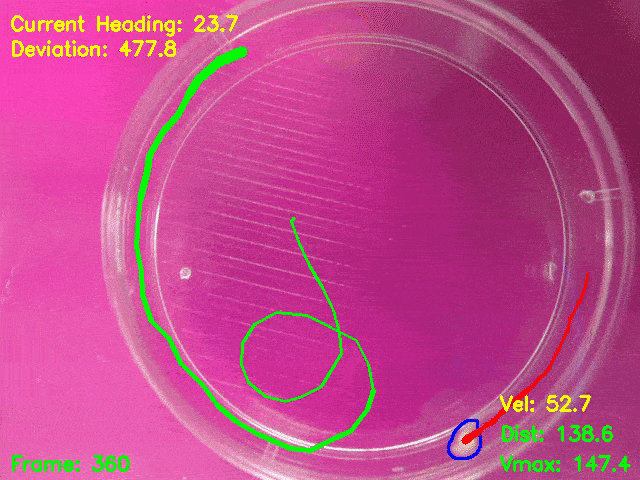
Automated tracking of a microglia-depleted tadpole making an escape response to bright looming stimuli.
Software available at https://github.com/tonykylim/XenLoom
-
CAD design and 3D printing
During the COVID-19 pandemic, nasopharyngeal swabs were in short supply. With a team that included 3D printing hobbyists, medical doctors, and bioengineers, we designed and developed a nasopharyngeal swab that could be made using low-cost FDM 3D printers, making it ideal for manufacturing in low resource settings.
Model available at https://3dprint.nih.gov/discover?terms=&uid=tonykylim
Learn more about this project at: https://www.helpfulengineering.org/projects-news/project-spotlight-open-source-3d-printed-nasopharyngeal-swab/
-
Bioinformatics

CysPresso, a tool that utilizes machine learning of deep-learning protein representations to predict the compatibility of therapeutic peptides for use in gene therapy.
This software was developed by Sébastien Ouellet.
Code is available at: https://github.com/Zebreu/cyspresso
Try it out yourself here:
https://colab.research.google.com/github/Zebreu/cyspresso/blob/main/CysPresso.ipynb
PUBLICATIONS

Peer-reviewed publications
Book chapters
AWARDS & FELLOWSHIPS

Awards
2023 Next Pharma Phenomenon team pitching competition, Department of Pharmacology, University of Cambridge
2018 Best Poster Award, EMBO Workshop: Microglia 2018
2015 1st Place Basic Science Poster Award, McGill Pain Day
2014 2nd Place Basic Science Poster Award, McGill Pain Day
2014 Oral Presentation Open Prize, McGill Anesthesia Research Day
2013 Poster Award, International Brain Barriers Society
2013 1st Place Oral Presentation, CIHR Neuroinflammation Training Program
2011 3rd Place Poster Presentation, CIHR Neuroinflammation Training Program
2011 Best Basic Science Poster Award, McGill Pain Day
2006 Esther R. Anderson Memorial Prize, UBC Department of Pharmacology
Fellowships
2022-2024
2017–2020
2016–2017
2012–2014
2009–2012
2006
Marie Skłodowska-Curie Actions European Postdoctoral Fellowship (UKRI Guarantee)
CIHR Postdoctoral Fellowship
McLaughlin McGill Faculty of Medicine Fellowship
The Edwards Foundation PhD Studentship in Pain Research
CIHR Frederick Banting and Charles Best Doctoral Fellowship
McGill Graduate Studies Fellowship
PRESENTATIONS

Selected oral presentations
“CysPresso: Predicting cysteine-dense peptide expression utilizing deep learning protein
representations.” PEGS Protein Engineering & Cell TherapySummit. Boston, MA. May 2024. (*Presented by co-author)
“Poster Highlight: Long-lasting expression of transgenes in mouse primary fibroblast-like
synoviocytes with self-amplifying RNA.” PEGS Protein Engineering & Cell Therapy
Summit. Boston, MA. May 2023.
“Microglial trogocytosis and the complement system regulate axonal pruning in vivo.” Canadian Neurophotonics Platform: Neuro Light Lunch Seminar Series, Virtual seminar, Jul 2020.
“In vivo imaging of microglial-mediated synaptic pruning in the Xenopus laevis retinotectal circuit and modulation by the complement system.” Xenopus Gene Editing Workshop, Woods Hole, MA, Oct 2019.
“In vivo imaging of microglial-mediated synaptic pruning in the developing retinotectal system.” Neuron-Glia Seminar Series, Montreal, Canada, Jun 2018.
“Do microglia eat synapses? An investigation by 2-photon live imaging.” University of Tokyo seminar, Tokyo, Japan, Mar 2018.
“Do microglia eat synapses? An investigation by 2-photon live imaging.” Kyoto University iCeMS seminar, Kyoto, Japan, Mar 2018.
“Do microglia eat synapses? An investigation by 2-photon live imaging.” Osaka University Frontier Bioscience Seminar, Osaka, Japan, Mar 2018.
“Do microglia eat synapses? Seeing is believing.” BRaIN Boost Symposium, Montreal, Canada, Feb 2018.
“The role of hypoxia in the generation of neuropathic pain.” Anesthesia Research Day, Montreal, Canada, May 2014.
“The vascular basis of neuropathic pain.” Quebec Network of Junior Pain Investigators, Montreal, Canada, Jul 2013.
“The vascular basis of neuropathic pain.” Integrated Program in Neuroscience Retreat, Montreal, Canada, Jun 2013
“A vascular hypothesis for neuropathic pain.” CIHR Neuroinflammation Training Program Symposium, Montreal, Canada, 2013.
RESEARCH EXPERIENCE

2022–2024
Research Associate
An 'immune-evasive' self-amplifying RNA platform for sustained and externally controllable transgene expression.
University of Cambridge
Cambridge, United Kingdom
Lab of Prof. Ewan St. John Smith
-
Engineered and characterized self-amplifying RNA constructs with reduced immunotoxicity for sustained and controllable transgene expression in mouse primary fibroblast-like synoviocytes.
2021–2022
Independent Scientist
CysPresso: a classification model utilizing deep learning protein representations to predict recombinant expression of cysteine-dense peptides
Unaffiliated
Vancouver, Canada
-
Led a bioinformatics project utilizing machine learning to predict the recombinant expression of cysteine-dense peptides in mammalian cells.
2016–2020
Postdoctoral Scholar
The role of microglial trogocytosis and the complement system in axonal pruning
The Montreal Neurological institute
Montreal, Canada
Lab of Dr Edward S. Ruthazer
-
Conducted in vivo studies using 2-photon microscopy to visualize microglial-mediated axonal
pruning in Xenopus laevis. -
Discovered an endogenous synapse-associated molecule that inhibits axonal pruning and engineered a synapse-associated fusion protein that enhances axonal pruning.
-
Developed a novel behavioral assay integrating programming, 3D printing, and computer vision for assessing visuomotor function.
2009–2015
Doctoral Student
The role of vascular dysfunction in neuropathic pain
McGill University
Montreal, Canada
Lab of Dr Ji Zhang
-
Investigated the response of peripheral nerve vasculature to nerve injury, focusing on endoneurial microvascular dysfunction, hypoxia, and metabolic disturbances.
-
Discovered three different mechanisms to target analgesics to injured peripheral nerves.
2007–2008
Master's Student
Evidence for a role of nerve injury in painful intervertebral disc degeneration: a cross-sectional proteomic analysis of human cerebrospinal fluid
McGill University
Montreal, Canada
Lab of Dr Laura S. Stone
-
Uncovered biomarkers that distinguish painful and asymptomatic phenotypes of degenerative disc disease in human cerebrospinal fluid using proteomics.
-
Uncovered biomarkers in the saliva of adults with self-injurious behaviour.
2006
Undergraduate Summer Research Project
The Quaternary Lidocaine Derivative, QX-314, Produces Long-lasting Local Anesthesia in Animal Models In Vivo
University of British Columbia
Vancouver, Canada
Labs of Dr Bernard A. MacLeod & Stephan K.W. Schwarz
-
Investigated the in vivo local anesthetic properties of the quaternary ammonium lidocaine derivative, QX-314.
2005-2006
Honour's Thesis Student
2005
Co-op Intern
2004
Co-op Intern
Summary of the discovery and development of RSD-921: From molecule to man
University of British Columbia
Vancouver, Canada
Lab of Dr Michael J.A. Walker
-
Summarized the discovery and development of RSD-921, a novel anti-arrhythmic.
Conventional and semi-high throughput electrophysiology of HCN ion channels
Johnson & Johnson Research & Development
La Jolla, San Diego, USA
Pain and Related Disorders Group
-
Performed conventional and high-throughput patch clamp electrophysiology on TRPV1, HCN, and hERG ion channels in primary rat dorsal root ganglion neurons and HEK293 cells.
Development of a clinically relevant rodent of allergic rhinitis
University of British Columbia
Vancouver, Canada
Supervisor: Michael J.A. Walker
-
Contributed to developing a guinea pig model of allergic rhinitis, facilitating in vivo drug screening.
EDUCATION

2009–2015
Doctor of Philosophy (PhD), Integrated Program in Neuroscience
2006–2008
Master of Science (MSc), Pharmacology & Therapeutics
2001–2006
Bachelor of Science (Hons),
Pharmacology & Therapeutics
McGill University
Thesis: The role of vascular dysfunction in neuropathic pain
McGill University
Thesis: Proteomic analysis of human cerebrospinal fluid from patients with painful and non-painful degenerative disc disease
The University of British Columbia
Thesis: A summary of the discovery and development of RSD-921, a novel anti-arrhythmic
TEACHING & MENTORING

Teaching
2023 - 2024
2023 - 2024
2023 - 2024
2022 - 2023
2022 - 2023
2022 - 2023
2019
Cambridge "supervisions" (small group tutorials)
Pharmacology NST Part IB
Laboratory practical demonstrator
Pharmacology NST Part IB
Laboratory practical demonstrator
Mechanisms of Drug Action MedST/VetST IB
Cambridge "supervisions" (small group tutorials)
Pharmacology NST Part II
Cambridge "supervisions" (small group tutorials)
Pharmacology NST Part IB
Laboratory practical demonstrator
Mechanisms of Drug Action MedST/VetST IB
Workshop: "Practical 3D printing for Life Scientists"
McGill University, Montreal Neurological Institute
-
Organized, created and presented a 3 hour workshop on 3D printing
Student supervision
2024
2020
2014
2013
2012
2011
T. Abdul (Undergraduate student, University of Cambridge)
J. Ho (Undergraduate student, McGill University)
J. Jang (Undergraduate student, McGill University)
J. Lou (Undergraduate student, University of Alberta)
J. Johnson (Undergraduate student, University of Alberta)
H. Martin (Undergraduate student, Memorial University)